Predicting polymer phase behaviour with machine learning
Predicting polymer phase behaviour is challenging and broadly relevant, yet the use of machine learning is constrained by inconsistent literature data that often lack critical parameters such as molecular weight, temperature, and concentration.
In this article, the authors highlight a study that leverages systematically collected experimental data with the Crystal16 instrument to develop a machine learning model for predicting the phase behavior of polymer solutions based on %transmission. The study examines 30 polymers across 45 unique solvents using a standardized experimental workflow, enabling evaluation of the model’s ability to generalize to previously unseen polymer–solvent combinations. Together, these results demonstrate how high-quality, standardized turbidity data can serve as a reliable foundation for data-driven prediction of polymer solubility behavior.

Experiment data generation
Solubility screening:
This was performed using the Crystal16 in a standardized workflow, with controlled temperature profiles from 10°C to 60°C, defined heating and cooling ramp rate of 0.5°C/min, and consistent stirring condition 0f 900 rpm applied over two heating–cooling cycles, including isothermal hold steps of 60 mins at 60°C, 120 mins at 10°C followed by final 60 mins hold at room temperature.
Crystal16 enables simultaneous measurement of up to 16 samples, allowing efficient and reproducible data collection under controlled conditions. Experiments were conducted mostly at one concentration of 15 mg/mL however more concentrations of 5, 15, 30, and 50 mg/mL were also included for some cases to strengthen model training and validation; concentration ranges selected for their relevance to polymer solution processing and formulation applications.
The polymers were chosen to span a diverse range of functional groups with most polymers falling below 15 kDa to minimize kinetic limitations and improve alignment with equilibrium behavior. Solvents were likewise selected to cover a broad polarity spectrum, including nonpolar, polar protic, and polar aprotic classes, ensuring wide chemical diversity within the dataset. The heatmap of the polymer-solvent pairs and their respected concentration is shown in Figure 1.
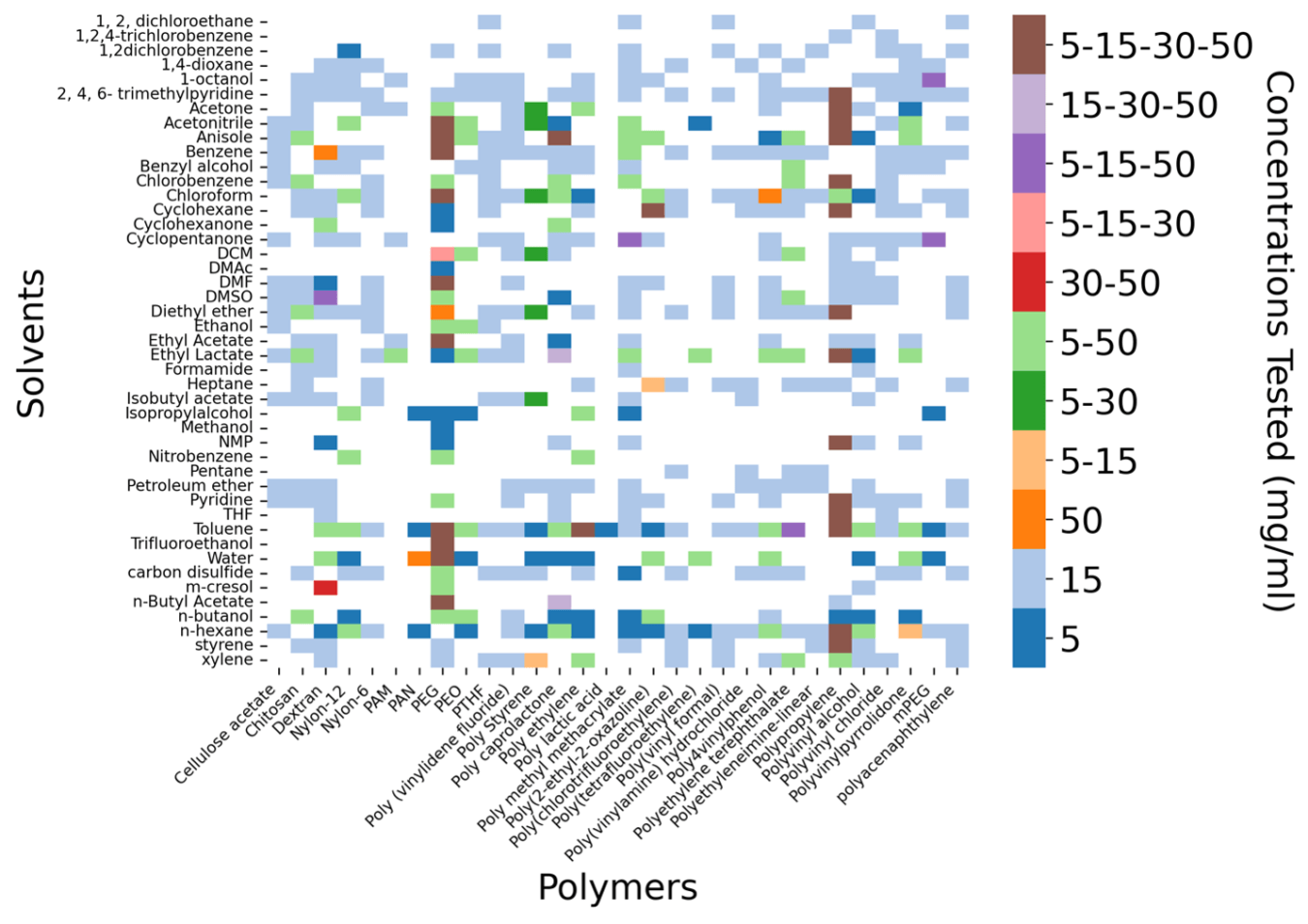
Figure 1. Heatmap of polymer-solvent pairs and their tested concentration using Crystal16. The color of each data cell indicates the concentration tested. brown denotes systems tested at four concentrations: 5, 15, 30, and 50 mg/mL.
Data Curation:
Crystal16 measurements generate transmission data as a function of temperature and time. Prior to modeling, the turbidity data were carefully curated to improve signal to noise ratio as some polymer solutions exhibit intrinsic noise and ensure consistency across experiments. To improve data consistency, the transmission profiles were selected during the cooling ramp and smoothed using a Savitzky–Golay filtering approach, allowing clearer transmission–temperature trends to be extracted for subsequent analysis. Out of the 880 reactors, 738 high-quality cooling-phase profiles comprising approximately 36,970 data points across different temperatures were selected.
Results:
The curated dataset was used to compare multiple solvent fingerprinting approaches, including Morgan, Kurotani, and Hansen descriptors, aimed at capturing solvent structural effects on polymer solubility. For polymers, a one-hot encoding scheme was applied to represent different polymer systems without introducing additional structural descriptors. These inputs were then used to train and evaluate random forest, XGBoost, and neural network models under a robust validation framework. One challenge associated with the dataset is that transmission values are heavily weighted toward 0% and 100%, reflecting systems that are either fully insoluble or fully soluble under the tested conditions. The final production model shown in Figure 2 was built using XGBoost with Morgan fingerprints and demonstrated strong predictive performance, achieving a root mean square error (RMSE) of approximately 6% and an R² of 0.98, indicating that it captures the majority of variability in the transmission data.
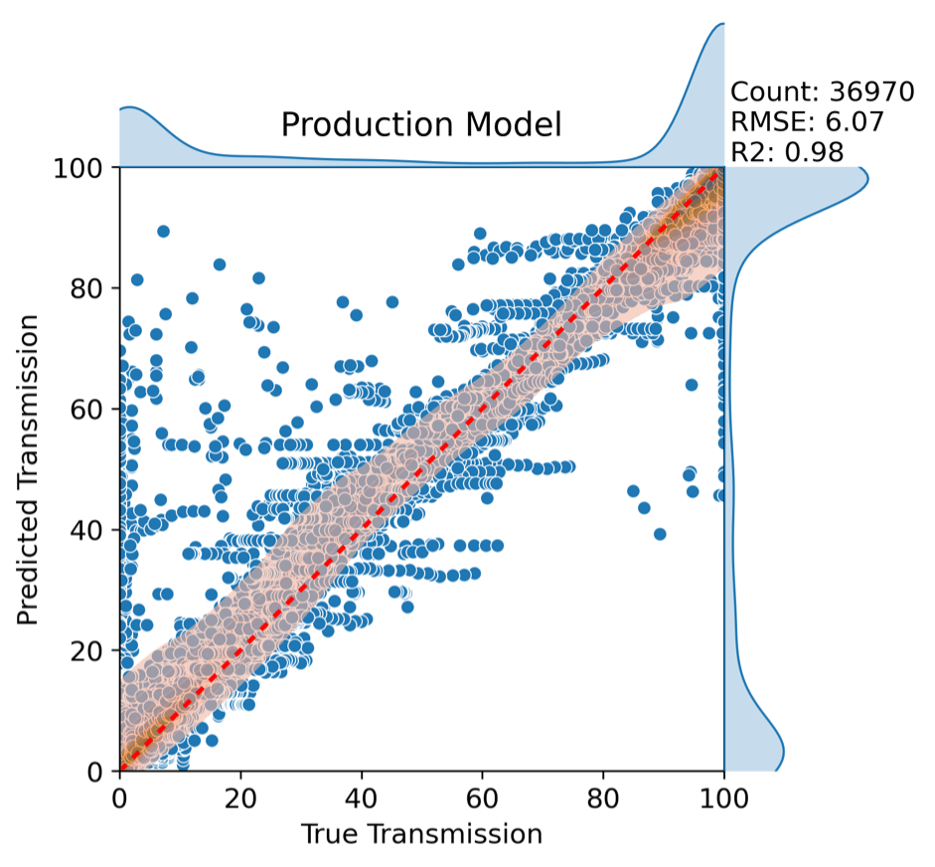
Figure 2: Predictions from the production XGBoost model trained with Morgan solvent fingerprints. The central two-dimensional kernel density estimate (KDE) highlights regions of highest data concentration, with darker orange indicating greater density. Marginal one-dimensional KDEs show the distributions of experimental transmission values (top) and model-predicted transmission values (right).
Model Performance Evaluation:
To assess how well the model performs on previously unseen data, a leave-one-out (LOO) analysis was conducted by systematically withholding individual polymer–solvent–concentration combinations during training. Model performance was first evaluated for completely new polymer–solvent pairs and then reassessed as increasing numbers of concentration data points for each pair were introduced. This approach highlights the model’s ability to generalize to new systems and demonstrates how predictive accuracy improves as additional concentration information becomes available, even with only a small number of measurements as shown in Figure 3.
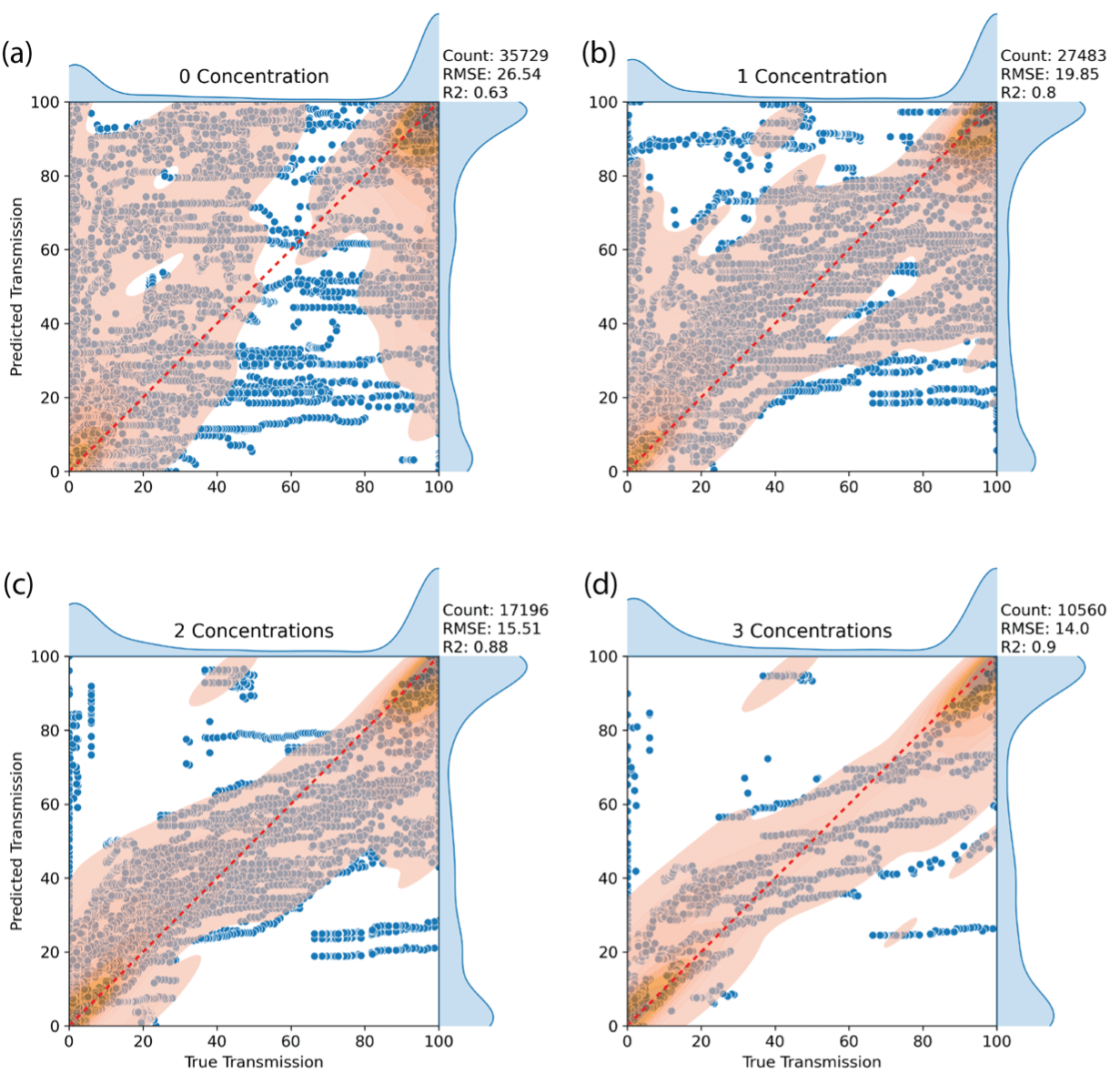
Figure 3: Leave-one-out parity plots for the XGBoost model using Morgan fingerprints, showing how predictive performance improves as increasing concentration data become available for each polymer–solvent pair.
Conclusion
This study highlights how high-quality, standardized turbidity data collected with the Crystal16 can be combined with machine learning to predict polymer solubility behavior. Using XGBoost with Morgan solvent fingerprints and one-hot polymer encoding, the model achieved strong predictive performance and demonstrated improved accuracy as additional concentration data collected with the Crystal16 were included, underscoring the value of well-controlled experimental datasets for data-driven polymer research.
References
We thank the authors for their valuable contributions and insights. Read the full article: https://pubs.acs.org/doi/10.1021/acs.jpcb.4c06500
Crystal16 for polymer studies
Curious how the instrument can advance your polymer research? Contact us for a demo tailored to your needs!